Most popular Neuroscience Blogs
I’m curious about what are the most popular blogs for neuroscience right now because I want to see what people are interested in, and learn how to write an interesting blog by example.
But how exactly do you find the most popular blog? There are blog ranking lists, top results on google, and lists of expert recommendations. Half the ones you find haven’t been updated in over a year.
Here I’ve analyzed some of these listings, and put my personal recommendations of up to date quality blogs in bold. (Updated June 2012.)

List I – Google Reader’s Top Results for Neuroscience Blogs or Neuro Blogs
1. Mind Hacks – Neuroscience and psychology tricks to find out what’s going on inside your brain. Features cool case studies, links to interesting articles, and analysis of the popular presses coverage of psychology issues. Most posts by Vaughan Bell, senior research fellow at the Institute of Psychiatry, King’s College London, with occassional offerings from Tom Stafford, Lecturer in Psychology and Cognitive Science in the Department of Psychology, University of Sheffield.
2. Action Potential – A Nature.com blog that is updated almost weekly. Similar in style to what I want to achieve, this blog lays out long summaries of papers recently published in nature, reports on conferences. Highly Reccomended. I really hope we start seeing more of this kind of blog.
Filed under: Blogs | 29 Comments
Tags: Blogs, Neuro Blogs, Neuroscience, Neuroscience Blogs
This article is my translation of this Brazilian article by RICARDO ZORZETTO (with lots of help from google translate)
Revising the Numbers
Get to know the anatomy of the human brain, especially how researchers got to a figure of 86 billion neurons,instead of the 100 billions previously estimated.
The complicatedly-named device is almost a meter high and resembles a garbage disposal. It contains electric motors that Continue reading ‘How many neurons do we really have? (And why we might have 10x fewer glia than we previously thought.)’
Filed under: Cerebellum, Comparative, Cortex, FACS - Fluorescent Automated Cell Sorting, Glia, Human, Primate, Rat | Leave a Comment
First, experimental economists and psychologists like nobel laureates Vernon L. Smith and Daniel Kahneman taught us that we aren’t economically rational–we’re influenced by biases and we use flawed heuristics (though often in very testable, repeatable ways).(1)
Then, Neuroeconomists showed that biology affects economic decisions–internasal oxytocin raises trust in risky exchanges, serum serotonin levels predict whether one will accept or reject an unfair offer from a stranger, and fMRIs demonstrate what brain systems are involved in our economic decision making.

Unfair offers in the ultimatum game conflict between emotional and cognitive more rational systems. (a) Areas in orange show greater activation following unfair as compared with fair offers–bilateral anterior insula and anterior cingulate cortex (ACC, right) and dorsolateral prefrontal cortex (dlPFC, left) (P<0.001). (b) Higher levels of right anterior insula activation predicts rejection of unfair offers. Sanfey (2006) Trends in Cognitive Sciences
Now, as The Boston Globe reports, genoeconomists are taking it further, confident they can use genetic variation to predict variation in financial behavior. (To which Princeton geneticist Leon Kruglyak tweeted “doubt it.”) Continue reading ‘Genoeconomics – Can genetic variation predict economic decisions?’
Filed under: Anatomy, Behavior, Dorsolateral PFC, Field / Technique, fMRI, Genetics, Genoeconomics, Human, Insula, Neuroeconomics, oxytocin, Peptides, Uncategorized, Vasopressin | Leave a Comment
Using machine learning, Harvard researchers create a web-based tool to diagnose autism in minutes
Original full text of the study available from Translational Psychiatry here.
Conventionally, children are diagnosed using Autism Diagnostic Interview, Revised (ADI-R) a 93-question survey, and/or the Autism Diagnostic Observation Schedule (ADOS) which measures behavior. The two test can take up to 2.5 hours and must be administered by clinical professionals. Dennis Wall–the lead author of the paper and director of the computational biology initiative at Harvard Medical School–says that “with the rising incidence of Autism parents often have to wait more than a year after initial warning signs and an appointment with a professional for official diagnoses,” and the earlier behavioral interventions start, the better.
Wall and colleagues set out to speed up diagnoses. Using machine learning analysis of ADI-R they found that only 7/93 questions were needed for near accuracy. Applying the same techniques to ADOS, they used an ADTree machine learning algorithm to shorten the test from 29 to 8 steps.

Schematic of the refined ADOS diagnostic classifier generated using ADTree machine learning algorithm. One follows each path originating from the top node and increments or decrements a prediction variable. At the end, if the variable is negative it indicates autism, with the magnitude corresponding to the confidence. Eight items were found to be significant:A2-Frequency of Vocalization Directed to Others, B1-Unusual Eye Contact, B2-Responsive Social Smile, B5-Shared Enjoyment in Interaction, B9-Showing, and B10-Spontaneous Initiation of Joint Attention, C1-Functional Play with Objects, and C2-Imagination / Creativity. Two items appeared more than once in the tree (B9 and B10) indicating they may be especially important for screening. Wall (2010) Translational Psychiatry
With the refined ADOS after training on one set of data, it classified two other sets of data, correctly diagnosing 334/336 individuals with autism, and with 94% specificity on a collection of observed and simulated non-spectrum controls.
Combined, the changes shorten the tests by nearly 95 percent–from around 2.5 hours to around 8 minutes. What’s more, Wall wants to put these tests online,so that the “caregiver will be able to take the crucial first steps to diagnosis and treatment from the comfort of their own home, and in just a few minutes.” Wall also asks the Autism community to help with the effort, by having the family member of someone clinically diagnosed with autism take an online survey or submit a short video to help verify and improve their tests.
Of course, autism diagnoses like most current psychiatric disorder diagnoses, are somewhat arbitrary in where they draw the line between Autism and Autism-like traits in neurotypical people. The algorithms may have to be adjusted next year with the release of the new psychiatric diagnostic manual, DSM 5.
Hopefully, these new tests will catch on within the psychiatric community increasing access to early diagnoses and early therapy when children’s brains are most plastic. Unfortunately the test only works for children two or older, but in the future functional imaging tests, eye-tracking, or genetic tests may allow earlier diagnoses.
———-
You may also be interested in my post: New sequencing study links autism to FMRP, the gene disrupted in fragile X syndrome and a regulator of synaptic plasticity
Filed under: Autism, Behavior, Disease, Human, Psychiatric, Uncategorized | 1 Comment
Scientists reported a new connection between autism and fragile x syndrome in the latest issue of neuron. They sequenced the exomes–the parts of DNA that code proteins–of 343 families that had a single child with autism and at least one unaffected sibling. Looking at de novo mutations (ones that occurred in the sperm or egg but not found throughout the parents), they found twice as many disruptive nonsense, splice site, and frame shift mutations in the sibling with autism as in controls (59 vs. 28). Rates of disruptive mutations were higher in females (9/29) versus males (50/314) p=.07, which matches up with data from other studies on gene copy number variations (CNVs) in autism. Levels of missense mutations appeared the same in autism and control groups, but it is often hard to judge whether a missense mutation will have a significant effect on protein function with extensive testing. Most mutations came from the fathers and rates increased with the age of the father.
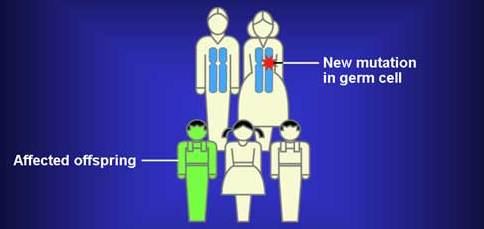
De Novo Mutations from http://www.cancer.gov/
Researchers compared the mutations they found to a previous study of gene copy number variations (CNVs) in autism. Two of the 59 disruptive mutations from the present study overlapped with the CNVs: Neurexin1 and PHF2.
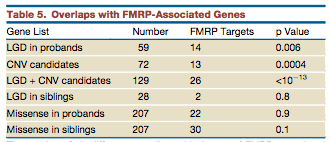
More interestingly, of the disruptive mutations in the autism group, 14/59 are part of the 842 genes regulated by FMRP (fragile X mental retardation gene product) (p=.006). Of the previously identified CNVs FMRP regulates 13/72 (p=.0004). In the disruptive mutations from controls, FMRP regulates only 2/28 a statistically insignificant amount.
When they analyzed the overall sample, and not just the de novo mutations, they also discovered an extreme scarcity of disruptive mutations in FMRP-associated genes. While they found 11.18% of synonymous variants falling within FMRP-associated genes (which would be roughly expected since they tend to be long genes), only .025% of the disruptive mutations fell into this category (P<10^-50). This strongly suggests that there is a purifying selection pressure against disruptive mutations to FMRP-associated genes.
Previously autism and FMRP had been connected because 90% of children with fragile X syndrome have autistic symptoms. FMRP regulates mRNA and is thought to inhibit synaptic protein translation of transcripts involved in synaptic plasticity, which is known to be essential for learning, memory, and general brain function, so from a mechanistic standpoint it is clear that such disruptions could impact cognition in atuism. Further, other regulators of synaptic plasticity such as TSC2 (tuberous sclerosis protein 2) and synaptic scaffolding proteins have also been implicated in autism.
The researchers hope to expand the study from the 343 families to 2800 families from the Simons Simplex Collection.
——
In additions to the advances we are making in the genetics of autism (and it appears to be one of the most heritable psychiatric disorders), we are making great progress in early diagnosis. Check out the post: Using machine learning, Harvard researchers create a web-based tool to diagnose autism in minutes.
You may also be interested in other articles about the neuroscience of psychiatric disorders.
Filed under: Autism, Disease, Fragile X, Genetics, Human, Plasticity, Psychiatric | Leave a Comment
Tags: ASD, de novo mutations, exome sequencing, FMRP, genetics of psychiatric disease, rare mutations, simons simplex, synaptic plasticity
Imagine cold fingers creeping up someone’s calf. Now imagine that whenever you saw someone else being touched, you would feel the sensation on your own body. That is mirror-touch synesthesia.
Psychologists at UCL verified mirror-touch synesthesia and further showed its linked with heightened empathy in their report in Nature Neuroscience.
Continue reading ‘Mirror Touch Synesthesia and the Genetics of Synesthesia’
Filed under: Behavior, Genetics, Human, SNPs, Uncategorized | 30 Comments
Can THC treat Anorexia and Bulemia? – Endocannabinoid systems are altered in eating disorders
“The munchies” — an effect of THC causing heightened craving and enjoyment of food after taking is currently used to help HIV patients and cancer patients undergoing chemotherapy patients maintain their weight. New research indicates that THC could help anorexics and bulemics regain weight–though probably not through “the munchies.”
A new PET imaging study reported in Biological Psychiatry suggests sufferers of anorexia and bulemia have a deficit in endocannabinoids (cannabinoids produced by the brain). Patients with eating disorders showed increased binding of a radio-labeled CB1R ligand [18F]MK-9470, and the authors of the study write that the upregulation of CB1R may be a compensation to a decrease in endocannabinoid signaling from the anorectic condition. CB1R Upregulation was especially notable in the insular cortex, which integrates interoception (stimuli produced from within, especially by the gut and other internal organs), taste, reward, and the processing of emotions such as disgust. Authors point out that the only clinical trial of cannabis on anorexia nervosa, high doses were used which can lower the appetite increasing effects.
However, authors caution that differences in body fat could confound the PET imaging of the lipophilic tracer. Also, the authors mention previous data conflicting with their hypothesis that the receptor upregulation may be due to endocannabinoid deficiency–studies have shown that endocannabinoid levels in the blood of anorexic patients are higher than in controls. However, it is unknown how well peripheral levels correlate with CNS levels, especially since endocannabinoids can have very local, temporally-specific effects. A better approximation than blood might be spinal tap samples of CSF, but that is quite a painful procedure. Even better might by in vivo magnetic resonance spectrometry for endocannabinoids in the insula. Can that be performed? Which chemicals can be detected with that method and which cannot?
—–
If you want to learn more about hunger, satiety, and weight lost check out my post on weight loss and longterm effects of hormones on hunger and satiety.
If you’re interested in the neuroscience of drugs check out: We know a lot about drugs – new data shows long-term epigenetic changes, and Card sorting, pot smoking, pleasure and one gene: COMT
Filed under: Drugs, Eating Disorders, Insula, PET scan, Pharmacology, Psychiatric | Leave a Comment
Tags: Anorexia, Bulemia, CB1, endocannabinoid, mk-9470, PET Study, THC
This two-day symposium, hosted by Drs. Ed Scolnick of the Broad Institute’s Stanley Center for Psychiatric Research, Li-Huei Tsai of MIT’s Picower Institute and Guoping Feng of MIT’s McGovern Institute, brought together leading scientists who work on the emerging genetics and biology of schizophrenia, bipolar disorder, autism and other mental illnesses. These illnesses cause lifelong disability in millions of people—combined, more than 3% of the world’s population is affected. We believe the timing of this symposium is ideal, as recent genetic findings are beginning to point to a new molecular understanding of the underlying pathophysiology as well as the commonalities and differences among these diseases. The four sessions reflect the flow of basic discoveries to new treatments, as the goal of this symposium is to reveal vast opportunities for new treatments for mental illnesses.
Topics include Genetics of Schizophrenia and Bipolar Illness, Genetics of Autism, OCD, Depression and Mental Retardation, Emerging Biology and Functional Data, and Clinical Applications.
Speaker List: Jonathan Sebat, Maria Karayiorgou, Michael O’Donovan, Shaun Purcell, Stephan Ripke, Pamela Sklar, Daniel Weinberger, Matthew State, Guoping Feng, Patrick Sullivan, Christopher A. Walsh, Ben Neale, Evan Eichler, Mario Capecchi, Elaine Lim, Mark Daly, Elly Nedivi, Li-Huei Tsai, Ed Boyden, Helen Mayberg, Rusty Gage, Jon Madison, Alea Mills, Peter Visscher, Jeff Conn, Ed Holson, Sasha Gragerov, Ronald Duman, Steve McKnight, Baltazar Gomez-Mancilla, Steve Hyman.
——
You may also be interested in my post on my favorite neuroscience podcasts: Beyond Blogs – Keep up with neuroscience through podcasts
Filed under: Genetics, Links, Psychiatric, Talks | Leave a Comment
Tags: Broad, GWAS, Mental Illness, Psychiatric, Sequencing, Stanley Center, Symposium
MPA, the Hormone used in the Depo Provera birth control shot, causes memory problems in rats
A new study published in Psychopharmacology shows young rats injected with medroxyprogesterone acetate (MPA) performed worse on behavioral memory tests. MPA’s memory impairment persisted even once it had been cleared from the blood.

MPA is the active component of the Depo Provera shot but not found in other hormonal contraceptives. The finding is especially significant because it is not true of all estrogens Continue reading ‘MPA, the Hormone used in the Depo Provera birth control shot, causes memory problems in rats’
Filed under: Behavior, Hippocampus, Hormones, Memory, Public Health, Rat | Leave a Comment
Tags: Depo Provera, GAD, MPA, Water Radial Arm Maze